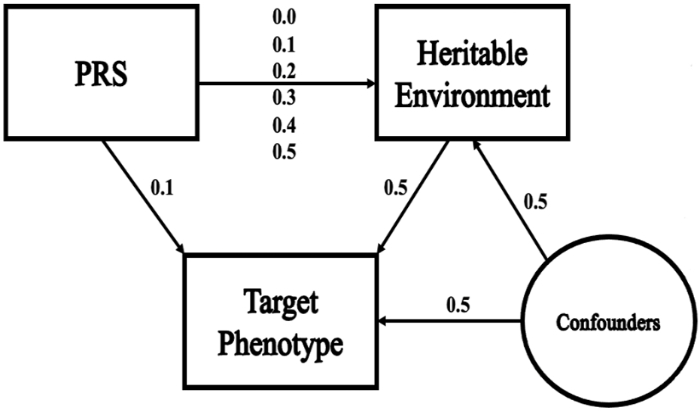
In this article, researchers explore an approach to correct collider bias persisting in existing cohort studies, specifically in genetic risk scoring for studying predispositions in certain study models. Genetic risk scoring is a widely used method to study predispositions to complex traits, including behaviors and psychiatric outcomes. However, statistical models, crafted to eliminate alternative explanations for associations between genetic risk scores and these traits, face potential bias if the alternate explanation is also influenced by the genetic risk score. Principal Component Analysis (PCA), a statistical approach, was used to leverage the wide variety of data that is available in many large cohort studies. Interestingly, when PCA is applied to real data from the Collaborative Study of the Genetics of Alcoholism (COGA), we found that using certain statistical factors helped reduce bias related to tobacco use. Specifically, it effectively diminished collider bias when tobacco use serves as the collider variable, potentially enhancing Polygenic Risk Score (PRS) effect size estimation and optimizing the use of available data resources in various studies. This approach makes efficient use of existing data resources in many large cohort studies, which should be treated as a high priority in complex trait genetics where data collection is costly and polygenic effect sizes are often small. In this regard, it can be used to make future estimations more accurate.
Thomas, N. S., Barr, P., Aliev, F., Stephenson, M., Kuo, S. I., Chan, G., Dick, D. M., Edenberg, H. J., Hesselbrock, V., Kamarajan, C., Kuperman, S., & Salvatore, J. E. (2022). Principal Component Analysis Reduces Collider Bias in Polygenic Score Effect Size Estimation. Behavior genetics, 52(4-5), 268–280. https://doi.org/10.1007/s10519-022-10104-z